When a deadly virus strikes, scientists must monitor its genome to help prevent its spread. University of Birmingham researcher Dr Josh Quick (PhD Microbiology, 2018) has been at the forefront of next-generation viral surveillance. His lab-in-a-suitcase was used to monitor and contain outbreaks of Ebola in Guinea and Zika in Brazil, and these techniques were also instrumental in enabling the worldwide genetic sequencing of COVID-19.
Genomic surveillance to contain a virus
Genomic science has advanced rapidly from when the human genome was sequenced for the first time only 20 years ago. Many new genomic applications are now routine including for inherited diseases and cancer treatment. One such development is called genomic surveillance, which is used to track how viruses spread. Traditionally used after an outbreak had taken place, Josh and his University of Birmingham team recognised the impact genomic sequencing could have on monitoring a virus as it unfolded in real-time.
He explains: 'When viruses replicate small changes, mutations are introduced which you can use to learn vital information about the virus, track transmission, monitor different variants and make sure vaccines remain efficient. This can help prevent the virus spreading, ultimately saving lives.'
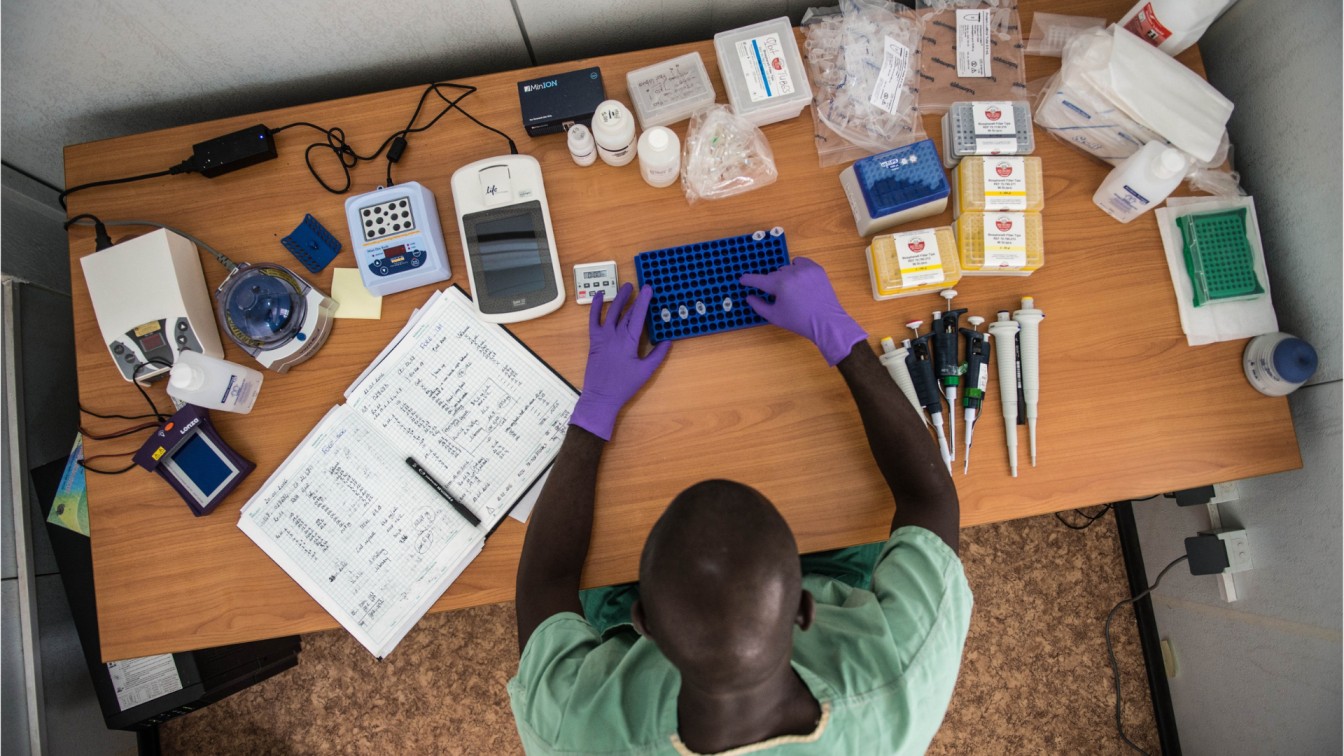
The "lab-in-a-suitcase"
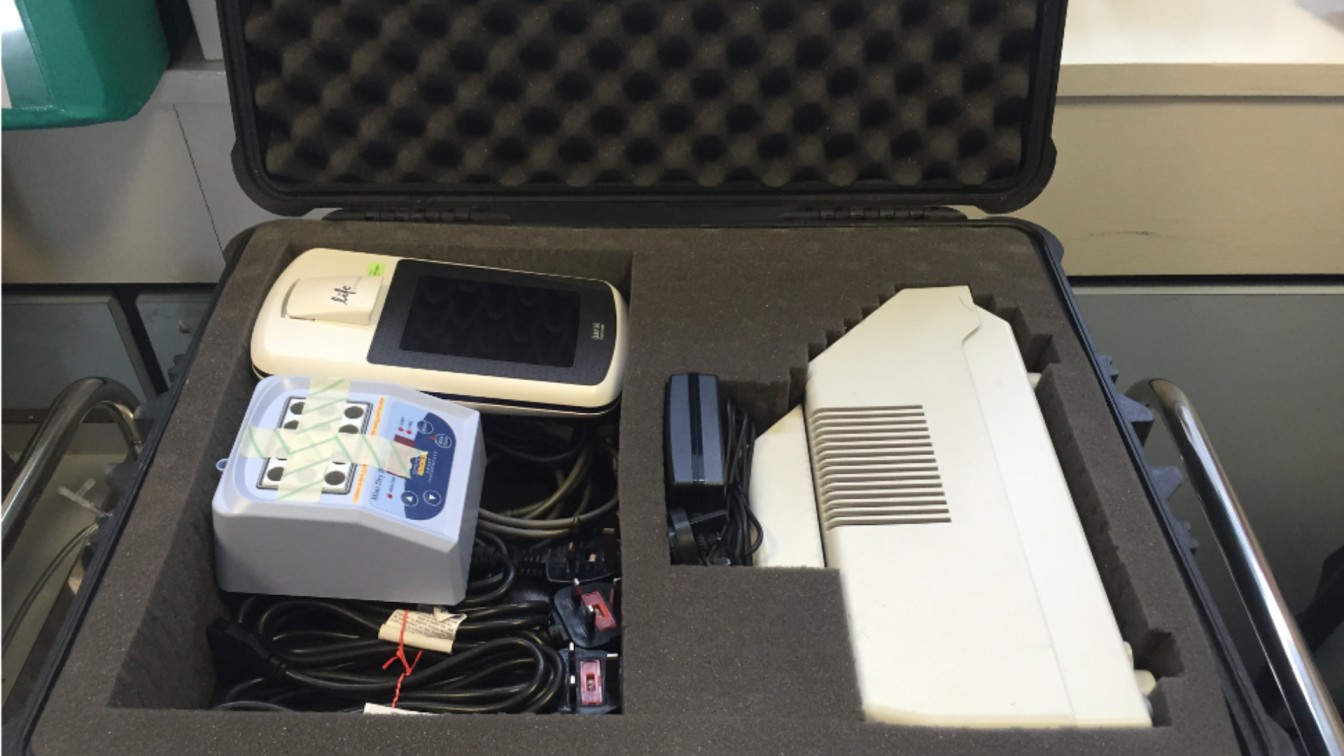
New technology meant Josh was able to compress his equipment to create a portable lab-in-a suitcase which could be taken on a plane as normal luggage, and enable a small sequencing lab to be set up anywhere in the world. He first used this technique during his PhD, travelling to Guinea in West Africa to establish a lab investigating the Ebola virus outbreak.
'Our idea was to use new Nanopore sequencing technology which allows real-time DNA analysis at the point of need. This enables huge amounts of information to be collected quickly to help inform health officials, doctors and public health specialists. Doing it on site meant we could sequence the virus quickly while the chain of transmission was still breakable. This was the start of what we now call real-time viral surveillance and it’s been applied to every major outbreak since.'
Lessons learnt from monitoring COVID-19
This experience cemented the team's status internationally, and when COVID-19 struck they were a vital part of viral surveillance through Genomics UK Consortium (COGUK). They were able to act incredibly quickly, creating a COVID-19 genome sequencing protocol within five days of the virus DNA being released. This was used by over 500 labs worldwide and was key to surveillance efforts and tracking the different variants worldwide. Josh says: 'We basically became a model system for how you do surveillance internationally. We were able to pick up the Kent variant first because of our expertise which was hugely significant. The network was also able to establish thousands of separate strands of the virus were initially introduced to the UK, a factor which could have huge implications for future public health policy.'
Applying it to Anti-Microbial Resistance (AMR)
Josh has recently applied his skills in viral surveillance to another huge threat, anti-microbial resistance, which happens when viruses and bacteria can no longer be treated by medicine. After gaining a prestigious UKRI Future Leaders Fellowship, he has been able to set up his own lab to focus on novel approaches to tackle the growing threat of resistance by rapidly identifying resistance genes in clinical samples.
Josh believes Nanopore sequencing has multiple possibilities for the future: 'This new technology opens up the possibility of sequencing for lots of different infections or pathogens that cause enormous mortality worldwide – the ultimate diagnostic test – in a cheap and accessible way.'
Find out more about Birmingham's research into infectious diseases.